Middle East Technical University, Departments of Electrical and Electronics Engineering, Neuroscience and Neurotechnology, and Health Sciences, Turkey
Authors:Ugur HALICI, Mustafa Ümit ÖNER, and Rengül Çetin Atalay
Abstract:The input of our method is whole slide images (WSIs) and outputs are metastasis region representatives together with their probabilities and whole slide probabilities. The method has mainly three steps, which are preprocessing, classification and post processing. Preprocessing was applied on Layer 7 images in order to determine lymph node sections and then only these sections were considered. A training set consisting of 64x64 RGB subimages from Layer 2 of original WSIs was constructed. Negative samples in the training set were selected from slides with label Normal and positive samples from metastasis regions of slides with label Tumor. A CNN having two convolutional layers for feature extraction and two fully connected layer and a softmax for classification was trained with these samples and used for classification of subimages of Layer 2 in the test set. After determining the labels of windowed image regions, we constructed two matrices: one for labels and the other for model outputs, i.e. class probability output of CNN for subimages in windows. In fact, label matrix is a binary image having the same size with Layer 7 images. Postprocessing consists of elimination of small regions and confidence filtering. Then metastasis region representatives were extracted for Evaluation 2 and whole slide probabilities were decided for Evaluation I.
Results:The following figure shows the receiver operating characteristic (ROC) curve of the method.
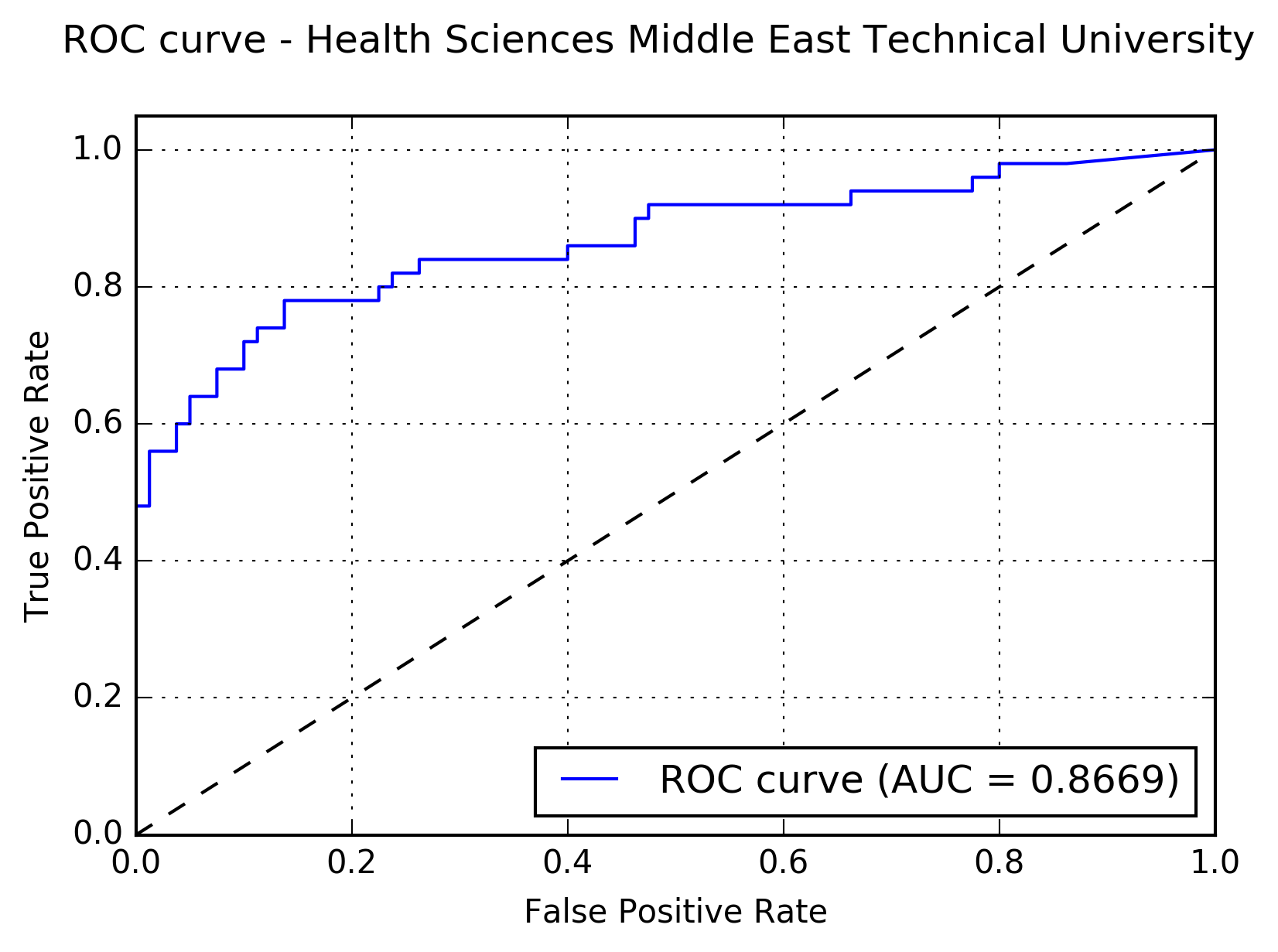
The following figure shows the free-response receiver operating characteristic (FROC) curve of the method.
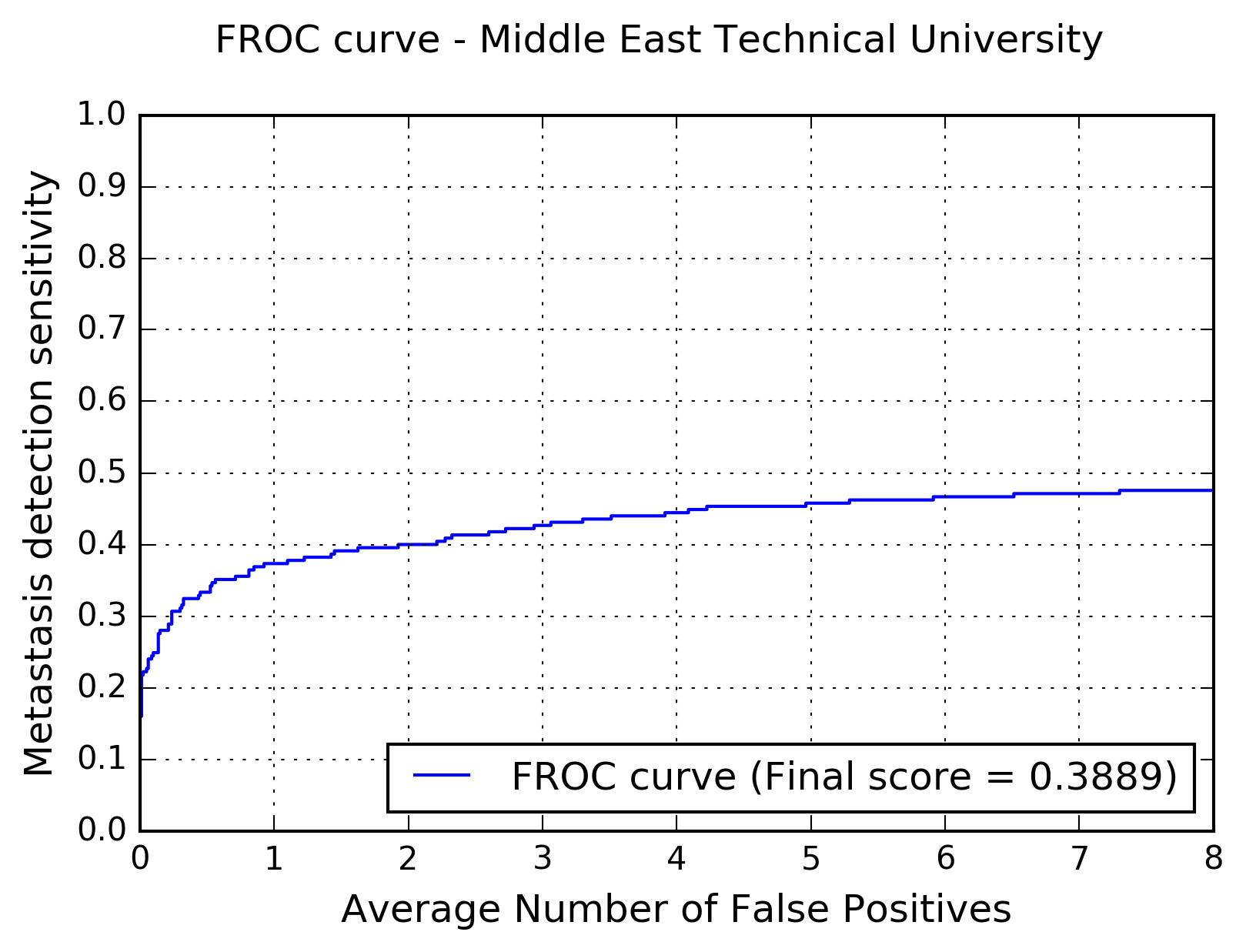
The table below presents the average sensitivity of the developed system at 6 predefined false positive rates: 1/4, 1/2, 1, 2, 4, and 8 FPs per whole slide image.
| FPs/WSI | 1/4 | 1/2 | 1 | 2 | 4 | 8 |
| Sensitivity | 0.307 | 0.333 | 0.373 | 0.400 | 0.444 | 0.476 |
