Osaka University, Department of Bioinformatic Engineering
Authors:Seiryo Watanabe , Shigeto Seno, Yoichi Takenaka, Hideo Matsuda
Abstract:We used Convolutional NeuralNetwork by GoogLeNet. At first, each slides has been subdivided into 300 square pixcels. Then, some of images were removed by file size and RGB value. We removed the size of less than 18000 bytes in jpeg format to ignore blank and scattered image. Then, we removed also less than 200 of Green and Blue value for same reason. We divided Normal, Marginal, Single and Tumor but changed to Normal and Tumor. Normal is from dataset of Train_Normal, Marginal have more than 50 percent region of Tumor, Single have one or more rounded tumor in the window and Tumor include entire. We trained with those data by using CNN and GoogLeNet, a 22 layers deep network. At first, we trained with those four group of images but changed to two group: Normal and Tumor which have Marginal and Single after several testings. Normal image dataset include 1,000,000 pieces and Tumor image dataset include 6,000 pieces. Training was performed as 66664 iteration and 2 epoch. The data has been convolved with filter of normalization as 1/9 * [1 1 1 ; 1 1 1 ; 1 1 1] and removed less than 0.1 for Fist Evaluation. Second Evaluation, we removed images less than 0.5.
Results:The following figure shows the receiver operating characteristic (ROC) curve of the method.
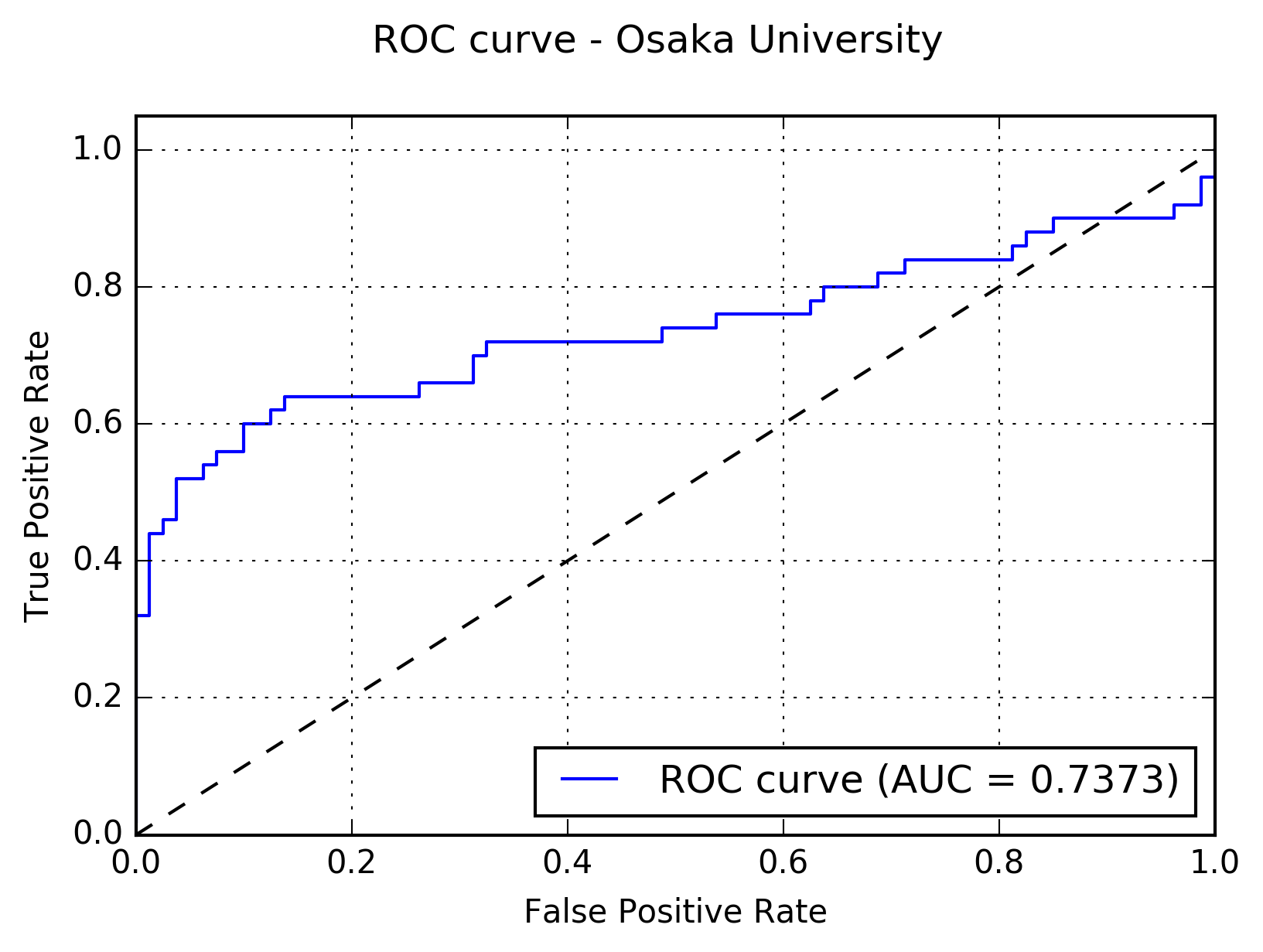
The following figure shows the free-response receiver operating characteristic (FROC) curve of the method.
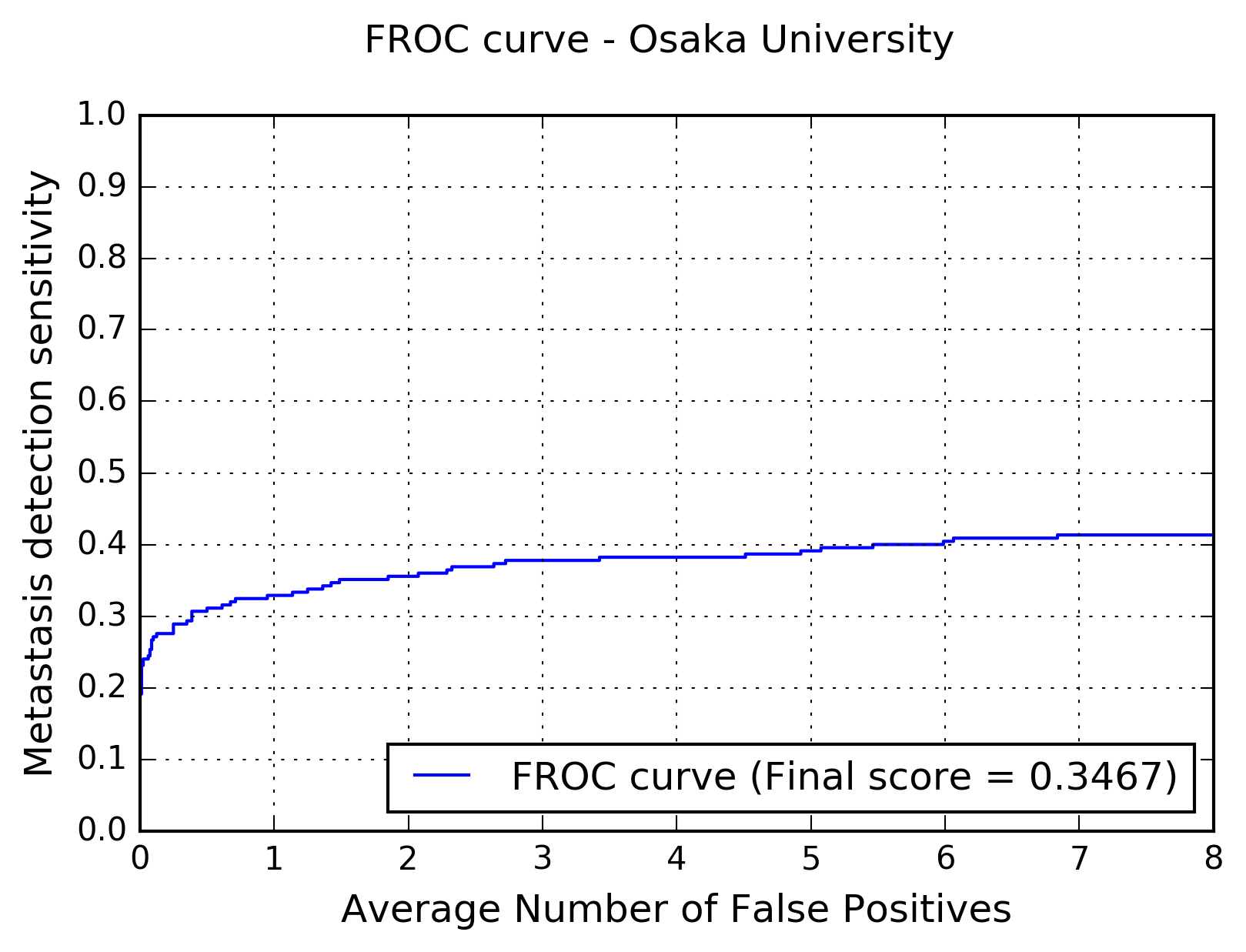
The table below presents the average sensitivity of the developed system at 6 predefined false positive rates: 1/4, 1/2, 1, 2, 4, and 8 FPs per whole slide image.
| FPs/WSI | 1/4 | 1/2 | 1 | 2 | 4 | 8 |
| Sensitivity | 0.289 | 0.311 | 0.329 | 0.356 | 0.382 | 0.413 |
