BioMediTech, University of Tampere, Finland
Authors:Mira Valkonen, Kimmo Kartasalo, Kaisa Liimatainen, Leena Latonen, Pekka Ruusuvuori
Abstract:Our method is based on feature engineering and supervised learning with random forest model. As a preprocessing step, color correction with histogram based method was done. The features extracted from the whole slide images include several local descriptors related to image morphology, texture, and spatial distribution of nuclei within the neighbourhood. As a result, each processed location was represented by a feature vector containing over 200 features. We used the feature representations of tissue samples to train a random forest model (an ensemble of 50 classification trees). The model used bootstrap aggregation to improve the stability and accuracy. The confidence values returned by the classifier were further processed using morphological operations and thresholded into binary detections. Distance from the boundaries of the detected area was used as a criterion for coordinate selection within the detected areas.
Results:The following figure shows the receiver operating characteristic (ROC) curve of the method.
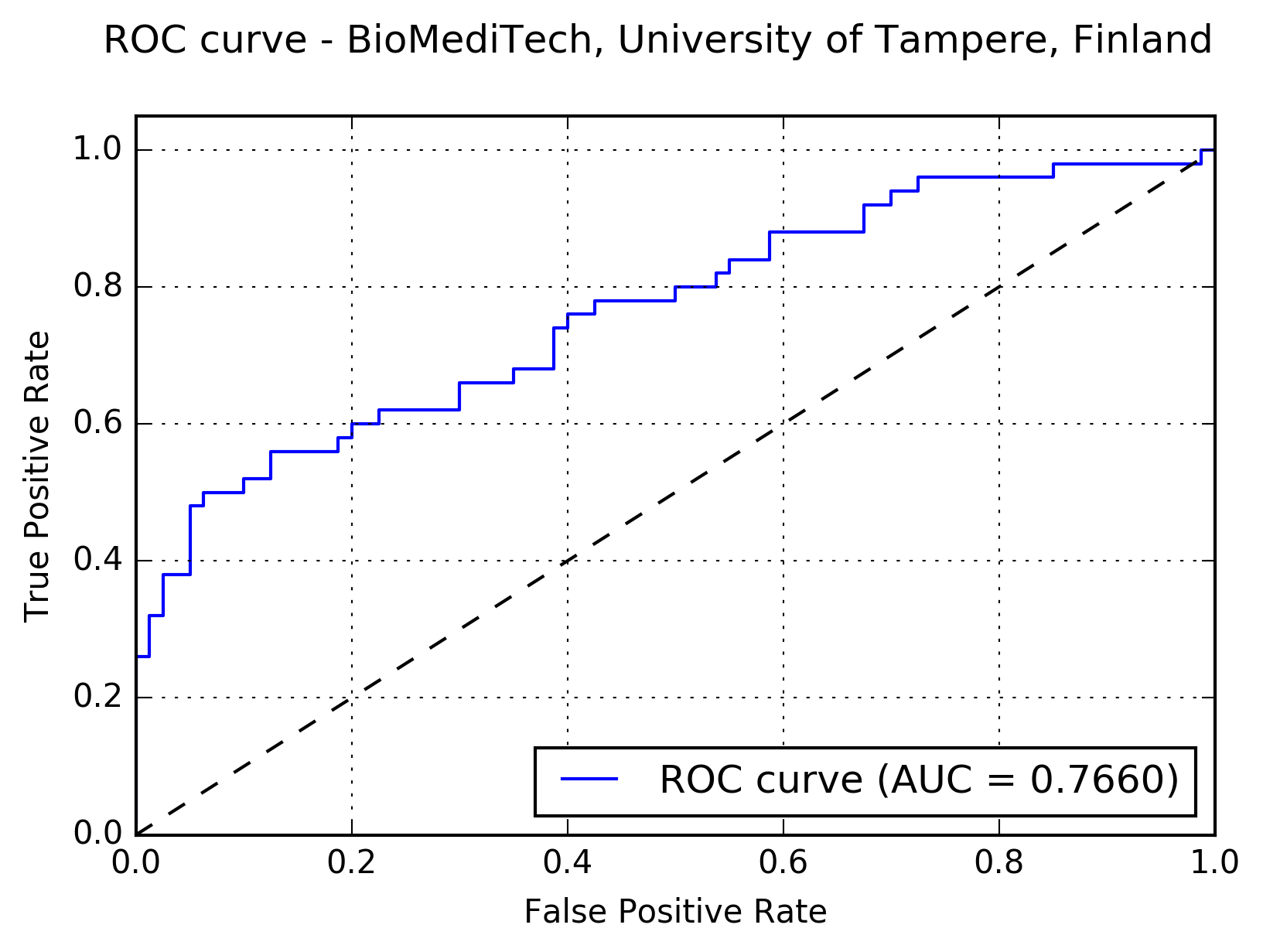
The following figure shows the free-response receiver operating characteristic (FROC) curve of the method.
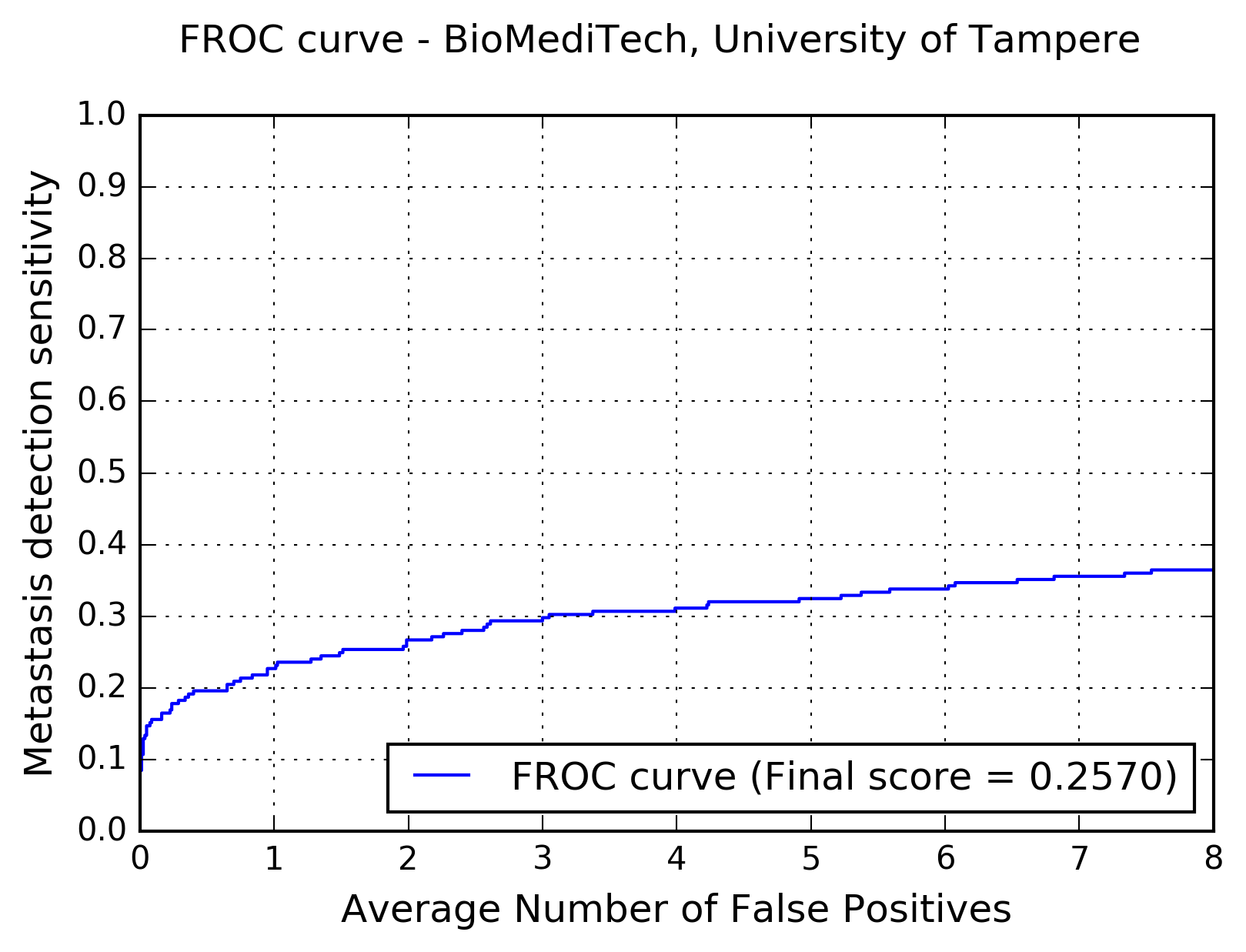
The table below presents the average sensitivity of the developed system at 6 predefined false positive rates: 1/4, 1/2, 1, 2, 4, and 8 FPs per whole slide image.
| FPs/WSI | 1/4 | 1/2 | 1 | 2 | 4 | 8 |
| Sensitivity | 0.178 | 0.196 | 0.227 | 0.267 | 0.311 | 0.364 |
