The Warwick-QU Team, Warwickshire, UK
Authors:Muhammad Shaban, Talha Qaiser, Ruqayya Awan, Korsuk Sirinukunwattana, Yee-Wah Tsang, and Nasir Rajpoot
Abstract:Our approach aims at segmenting the tumor regions by using a variant of the U-Net convolutional-deconvolutional neural network as the main component. Before feeding image patches to the network, we first performed background removal and stain normalization to deal with stain variation in the H&E stained slide images. We segmented the tissue region (area of interest) from fatty and white background areas by using a simple fully convolutional network (FCN) with a single upsampling layer. We then implemented the architecture of U-Net that is customised for the task of tumor segmentation in many ways. Our implementation in TensorFlow allows ease of customisation. The output of the network is a probability map, whereby each pixel intensity value corresponds to the probability of it belonging to the tumor, which is further manipulated by taking into account the probability values within a region and the region area. The candidate regions are further refined using the weighted probability map. For training purposes, we collected 20,000 RGB patches of size 428X428 at magnification level 2 (10X), with 12,000 patches taken from normal WSIs and 8,000 patches from WSIs with metastasis. The training and validation datasets comprised of 90% and 10% of the whole dataset respectively and the network was trained for more than 50 epochs.
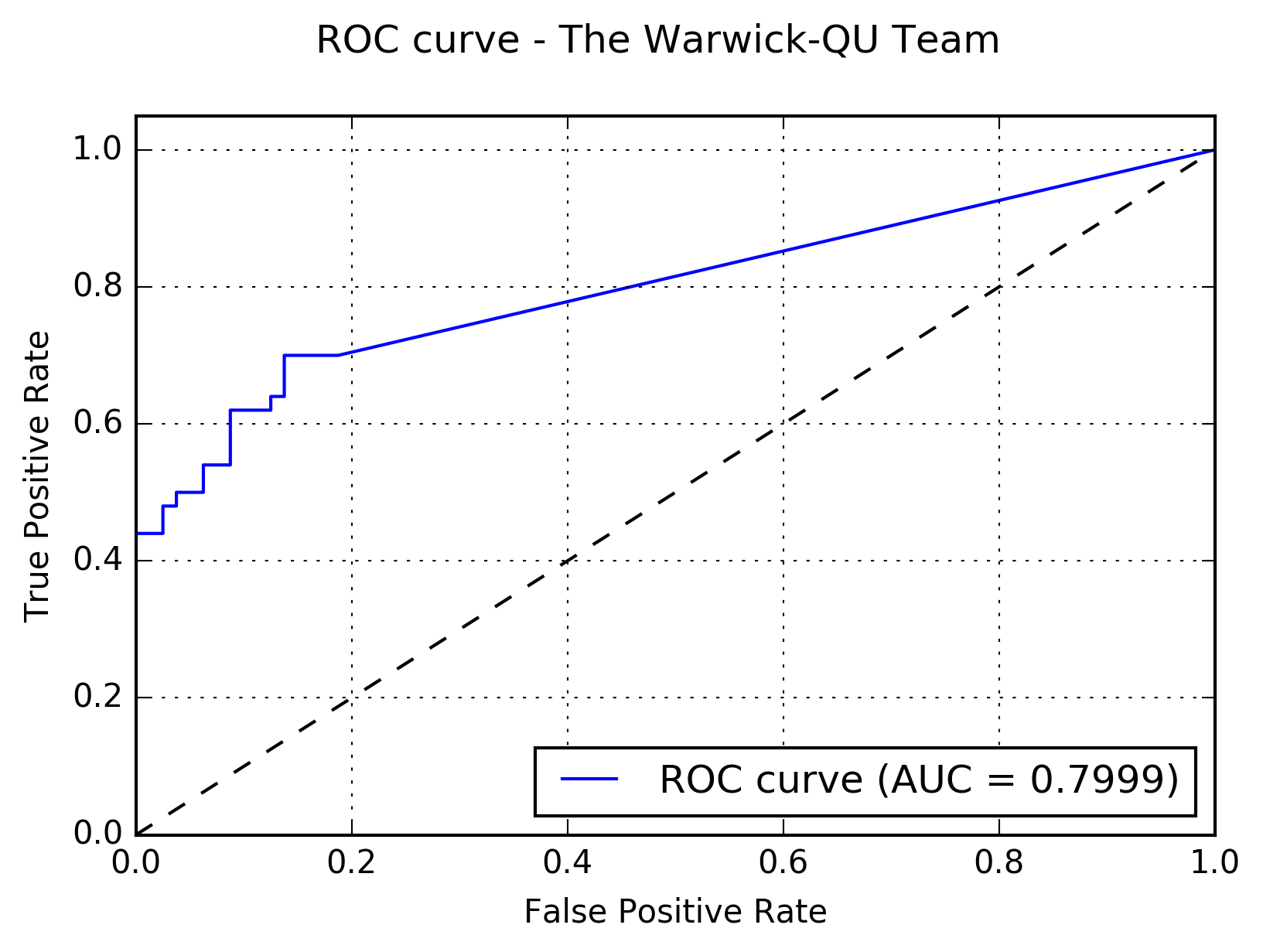
Results:The following figure shows the receiver operating characteristic (ROC) curve of the method.
The following figure shows the free-response receiver operating characteristic (FROC) curve of the method.
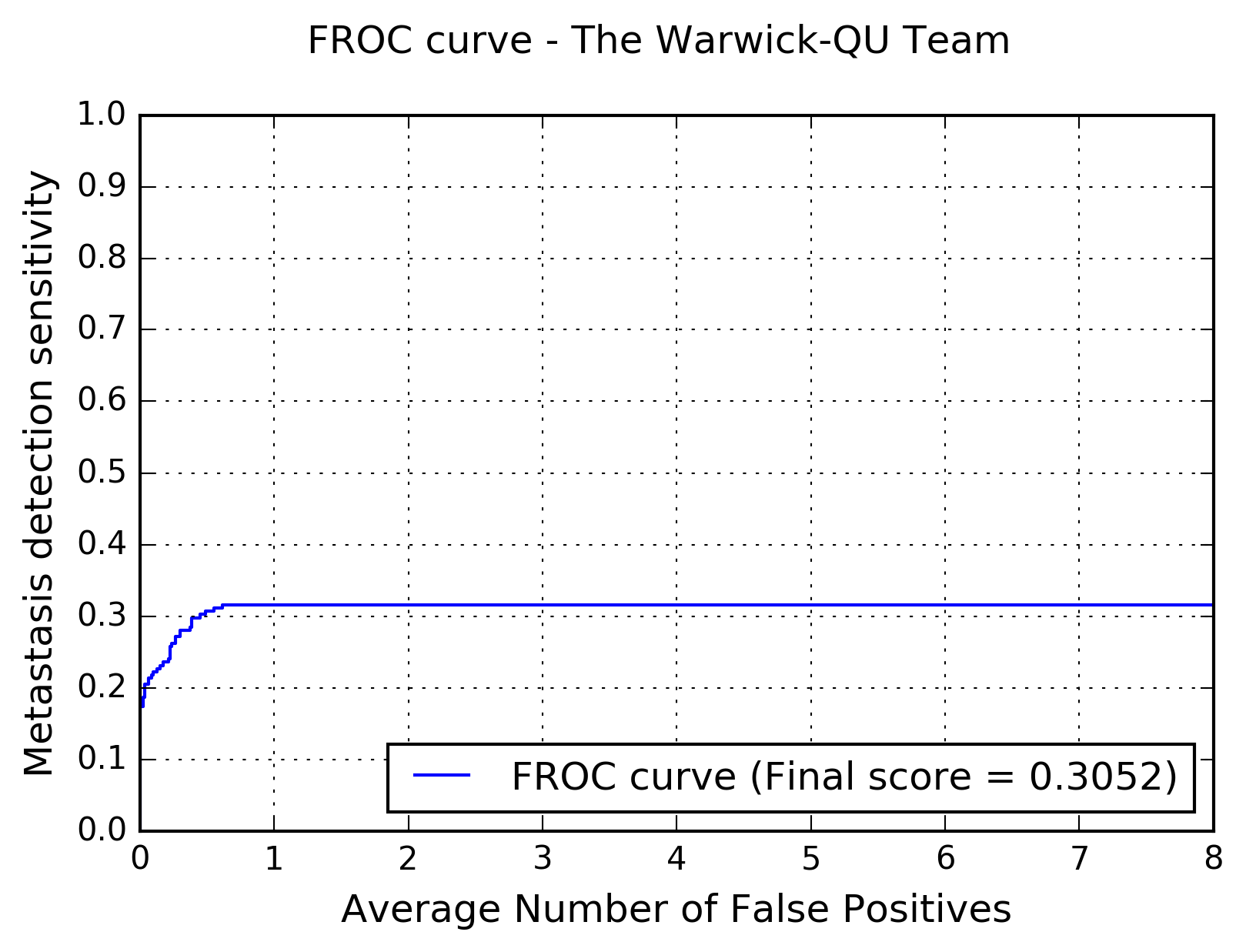
The table below presents the average sensitivity of the developed system at 6 predefined false positive rates: 1/4, 1/2, 1, 2, 4, and 8 FPs per whole slide image.
| FPs/WSI | 1/4 | 1/2 | 1 | 2 | 4 | 8 |
| Sensitivity | 0.262 | 0.307 | 0.316 | 0.316 | 0.316 | 0.316 |
